Back to all members...
Pascal Notin
PhD, started 2019

Pascal is a DPhil student in the CS Department at Oxford University, supervised by Yarin Gal. His research interests lie at the intersection of Bayesian Deep Learning, Generative Models, Causal Inference and Computational Biology. The current focus of his work is to develop methods to quantify and leverage uncertainty in models for structured representations (e.g., sequences, graphs), with applications in biology and medicine.
He has several years of applied machine learning experience developing AI solutions, primarily within the healthcare and pharmaceutical industries (e.g., Real World Evidence, treatment adherence, disease prediction). Prior to coming to Oxford, he was a Senior Manager at McKinsey & Company in the New York and Paris offices, where he was leading cross-disciplinary teams on fast-paced analytics engagements.
He obtained a M.S. in Operations Research from Columbia University, and a B.S. and M.S. in Applied Mathematics from Ecole Polytechnique. He is a recipient of a GSK iCASE scholarship.
News items mentioning Pascal Notin • Publications while at OATML • Reproducibility and Code • Blog Posts
News items mentioning Pascal Notin:

OATML researchers publish paper in Nature
03 Nov 2021
The paper “Disease variant prediction with deep generative models of evolutionary data” was published in Nature. The work is a collaboration between OATML and the Marks lab at Harvard Medical School. It was led by Pascal Notin and Professor Yarin Gal from OATML together with Jonathan Frazer, Mafalda Dias, and Debbie Marks from the Marks lab. OATML DPhil student Aidan Gomez and Marks lab researchers Joseph Min and Kelly Brock are co-authors on the paper.

NeurIPS 2021
11 Oct 2021
Thirteen papers with OATML members accepted to NeurIPS 2021 main conference. More information in our blog post.

OATML graduate students receive best reviewer awards and serve as expert reviewers at ICML 2021
06 Sep 2021
OATML graduate students Sebastian Farquhar and Jannik Kossen receive best reviewer awards (top 10%) at ICML 2021. Further, OATML graduate students Tim G. J. Rudner, Pascal Notin, Panagiotis Tigas, and Binxin Ru have served the conference as expert reviewers.

OATML student receives a best poster award at the ICML workshop on Computational Biology
07 Aug 2021
OATML MSc student Lood van Niekerk received a best poster award at the 2021 ICML Workshop on Computational Biology for his work on “Exploring the latent space of deep generative models: Applications to G-protein coupled receptors”. This is part of an ongoing collaboration between OATML and the Marks Lab. From OATML, Professor Yarin Gal and DPhil student Pascal Notin are co-authors on the paper.

ICML 2021
17 Jul 2021
Seven papers with OATML members accepted to ICML 2021, together with 14 workshop papers. More information in our blog post.

OATML graduate students receive Outstanding Reviewer Awards
03 Jun 2021
OATML graduate students Pascal Notin and Tim G. J. Rudner received best reviewer awards (top 5%) at UAI 2021.

OATML student invited to speak at the Cornell ML in Medicine seminar series
19 Mar 2021
OATML graduate student Pascal Notin will give an invited talk on Uncertainty in deep generative models with applications to genomics and drug design at the Cornell Machine Learning in Medicine seminar series. Professor Yarin Gal and collaborator José Miguel Hernández-Lobato are co-authors on the paper.

OATML student invited to speak at the EMBL workshop on ML in Drug Discovery
11 Mar 2021
OATML graduate student Pascal Notin will give an invited talk on “Large-scale clinical interpretation of genetic variants using evolutionary data and deep generative models” at the EMBL-EBI workshop on Machine Learning in Drug Discovery. The work is a collaboration between OATML (Pascal and Professor Yarin Gal) and the Marks Lab at Harvard.
Publications while at OATML:
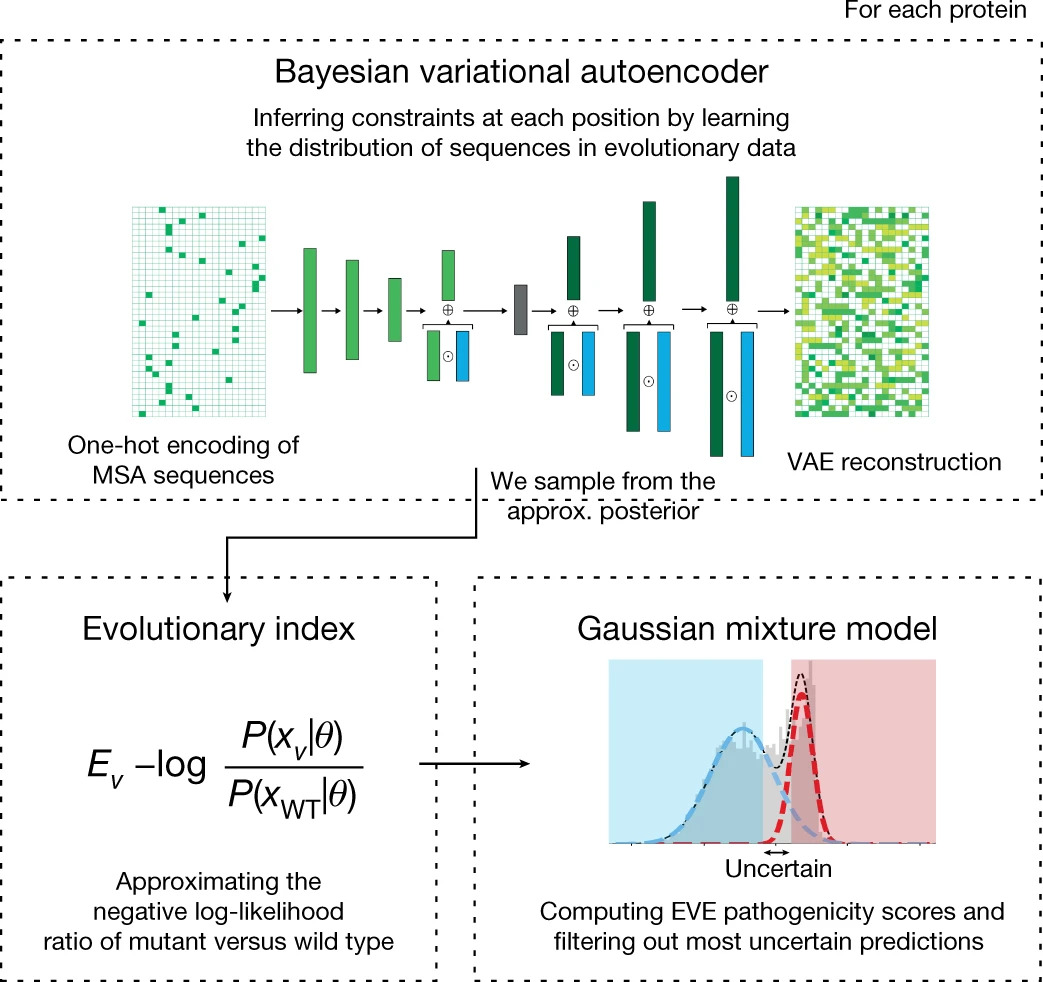
Disease variant prediction with deep generative models of evolutionary data
Quantifying the pathogenicity of protein variants in human disease-related genes would have a marked effect on clinical decisions, yet the overwhelming majority (over 98%) of these variants still have unknown consequences. In principle, computational methods could support the large-scale interpretation of genetic variants. However, state-of-the-art methods have relied on training machine learning models on known disease labels. As these labels are sparse, biased and of variable quality, the resulting models have been considered insufficiently reliable. Here we propose an approach that leverages deep generative models to predict variant pathogenicity without relying on labels. By modelling the distribution of sequence variation across organisms, we implicitly capture constraints on the protein sequences that maintain fitness. Our model EVE (evolutionary model of variant effect) not only outperforms computational approaches that rely on labelled data but also performs on par with, if... [full abstract]
Jonathan Frazer, Pascal Notin, Mafalda Dias, Aidan Gomez, Joseph K. Min, Kelly Brock, Yarin Gal, Debora Marks
[Nature]
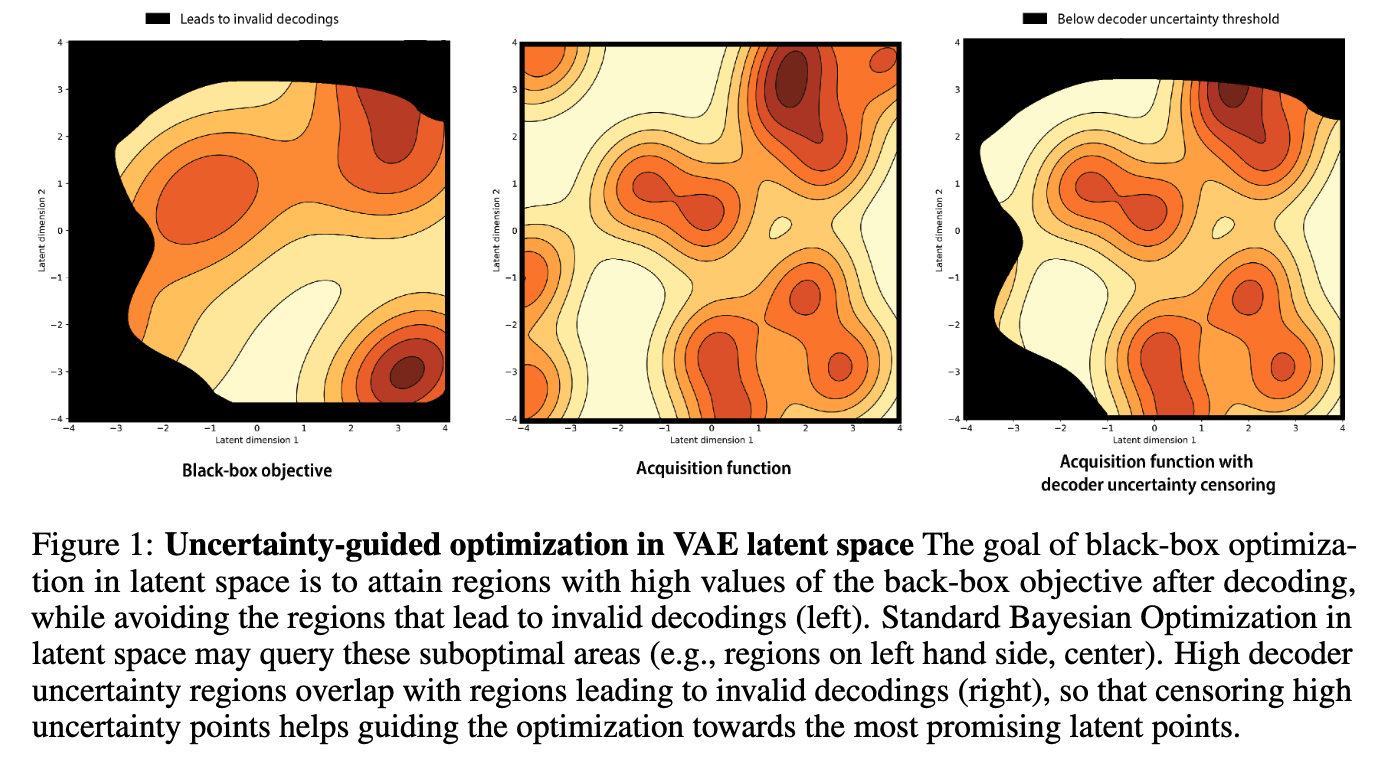
Improving black-box optimization in VAE latent space using decoder uncertainty
Optimization in the latent space of variational autoencoders is a promising approach to generate high-dimensional discrete objects that maximize an expensive black-box property (e.g., drug-likeness in molecular generation, function approximation with arithmetic expressions). However, existing methods lack robustness as they may decide to explore areas of the latent space for which no data was available during training and where the decoder can be unreliable, leading to the generation of unrealistic or invalid objects. We propose to leverage the epistemic uncertainty of the decoder to guide the optimization process. This is not trivial though, as a naive estimation of uncertainty in the high-dimensional and structured settings we consider would result in high estimator variance. To solve this problem, we introduce an importance sampling-based estimator that provides more robust estimates of epistemic uncertainty. Our uncertainty-guided optimization approach does not require modifica... [full abstract]
Pascal Notin, José Miguel Hernández-Lobato, Yarin Gal
Principled Uncertainty Estimation for High Dimensional Data
The ability to quantify the uncertainty in the prediction of a Bayesian deep learning model has significant practical implications—from more robust machine-learning based systems to more effective expert-in-the loop processes. While several general measures of model uncertainty exist, they are often intractable in practice when dealing with high dimensional data such as long sequences. Instead, researchers often resort to ad hoc approaches or to introducing independence assumptions to make computation tractable. We introduce a principled approach to estimate uncertainty in high dimensions that circumvents these challenges, and demonstrate its benefits in de novo molecular design.
Pascal Notin, José Miguel Hernández-Lobato, Yarin Gal
Uncertainty & Robustness in Deep Learning Workshop, ICML, 2020
[Paper]
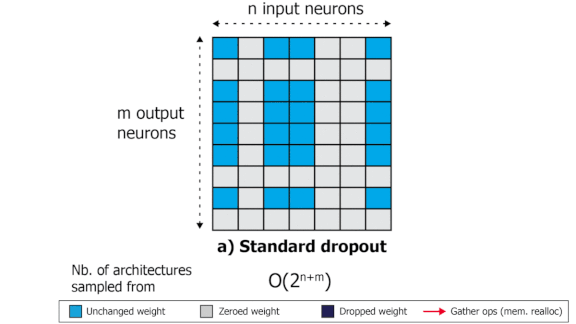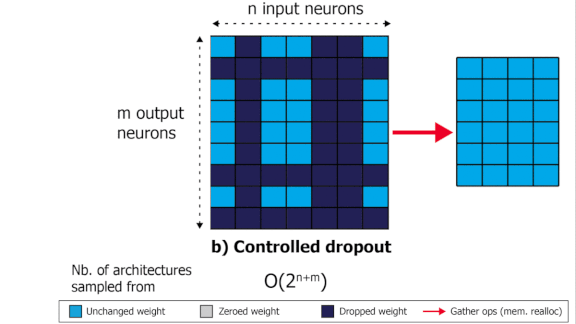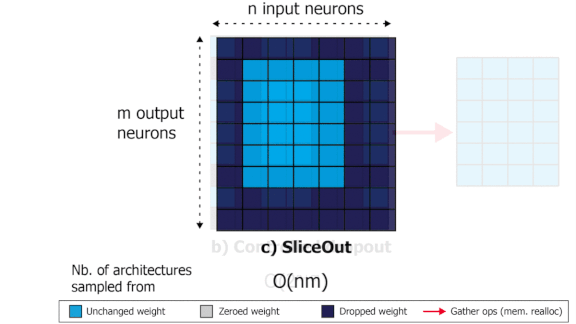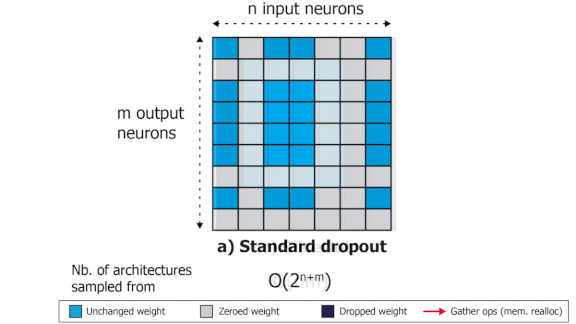
SliceOut: Training Transformers and CNNs faster while using less memory
We demonstrate 10-40% speedups and memory reduction with Wide ResNets, EfficientNets, and Transformer models, with minimal to no loss in accuracy, using SliceOut---a new dropout scheme designed to take advantage of GPU memory layout. By dropping contiguous sets of units at random, our method preserves the regularization properties of dropout while allowing for more efficient low-level implementation, resulting in training speedups through (1) fast memory access and matrix multiplication of smaller tensors, and (2) memory savings by avoiding allocating memory to zero units in weight gradients and activations. Despite its simplicity, our method is highly effective. We demonstrate its efficacy at scale with Wide ResNets & EfficientNets on CIFAR10/100 and ImageNet, as well as Transformers on the LM1B dataset. These speedups and memory savings in training can lead to CO2 emissions reduction of up to 40% for training large models.
Pascal Notin, Aidan Gomez, Joanna Yoo, Yarin Gal
Under review
[Paper]
Reproducibility and Code
Disease variant prediction with deep generative models of evolutionary data
Official repository for our paper “Disease variant prediction with deep generative models of evolutionary data”. The code provides all functionalities required to train the different models leveraged in EVE (Evolutionary model of Variant Effect), as well as the ones to score any mutated protein sequence of interest.
CodePascal Notin
Blog Posts
13 OATML Conference papers at NeurIPS 2021
OATML group members and collaborators are proud to present 13 papers at NeurIPS 2021 main conference. …
Full post...Jannik Kossen, Neil Band, Aidan Gomez, Clare Lyle, Tim G. J. Rudner, Yarin Gal, Binxin (Robin) Ru, Clare Lyle, Lisa Schut, Atılım Güneş Baydin, Tim G. J. Rudner, Andrew Jesson, Panagiotis Tigas, Joost van Amersfoort, Andreas Kirsch, Pascal Notin, Angelos Filos, 11 Oct 2021
21 OATML Conference and Workshop papers at ICML 2021
OATML group members and collaborators are proud to present 21 papers at ICML 2021, including 7 papers at the main conference and 14 papers at various workshops. Group members will also be giving invited talks and participate in panel discussions at the workshops. …
Full post...Angelos Filos, Clare Lyle, Jannik Kossen, Sebastian Farquhar, Tom Rainforth, Andrew Jesson, Sören Mindermann, Tim G. J. Rudner, Oscar Key, Binxin (Robin) Ru, Pascal Notin, Panagiotis Tigas, Andreas Kirsch, Jishnu Mukhoti, Joost van Amersfoort, Lisa Schut, Muhammed Razzak, Aidan Gomez, Jan Brauner, Yarin Gal, 17 Jul 2021
13 OATML Conference and Workshop papers at ICML 2020
We are glad to share the following 13 papers by OATML authors and collaborators to be presented at this ICML conference and workshops …
Full post...Angelos Filos, Sebastian Farquhar, Tim G. J. Rudner, Lewis Smith, Lisa Schut, Tom Rainforth, Panagiotis Tigas, Pascal Notin, Andreas Kirsch, Clare Lyle, Joost van Amersfoort, Jishnu Mukhoti, Yarin Gal, 10 Jul 2020