Irina Voiculescu

Professor Irina Voiculescu
Themes:
Completed Projects:
Interests
My research develops machine learning methods for medical image analysis, including segmentation, 3D reconstruction, and anatomical landmark detection. Current projects integrate multi-modality data to improve diagnostic accuracy, while ensuring trust and fairness in healthcare AI.
Research
I lead the Oxford Medical Image Science (OxMedIS) group, which works across x-ray, MRI, CT, ultrasound, and other imaging data. We design algorithms that detect and classify clinically relevant features, reconstruct anatomical structures in 3D, and provide tools that balance automation with clinician input. Beyond image analysis, we address failure detection and model calibration to ensure reliability, with the ultimate goal of translating these methods into clinical practice.
Vacancies
Postdoctoral positions are advertised on the list of vacancies. Interdisciplinary postoctoral applicants from areas other than Computer Science may check their eligibility for the Schmidt AI Fellowships.
Prospective DPhil/PhD candidates who meet the admissions eligibility criteria are most welcome to apply to join my group. Students taking the MSc, MCompSci and related courses can work with us by choosing or proposing a project in the topics below. Regrettably we cannot accommodate internships.
Our main clinical collaborators specialize in orthopaedics, which constitutes our primary data source. Proposals for research projects for a different clinical field are gladly received, but please state your expected data provenance, or how we can work together to secure an appropriate dataset.
Prior to applying for a graduate place, please read the information already available online, and get in touch informally describing your interest in
- Segmentation from 2D images or 3D volumes
- Small structure localisation from weak annotations (scribbles, dots)
- 3D reconstruction of anatomical structures
- Anatomical landmark detection
- Validation and evaluation measures (in the presence of multiple annotators)
- Fairness and bias detection in healthcare
- Failure detection in image classification
- Multi-modality data integration for more accurate diagnosis
- Trust and explainability in clinical applications
Clinical applications, using different data acquisition modalities
X-ray
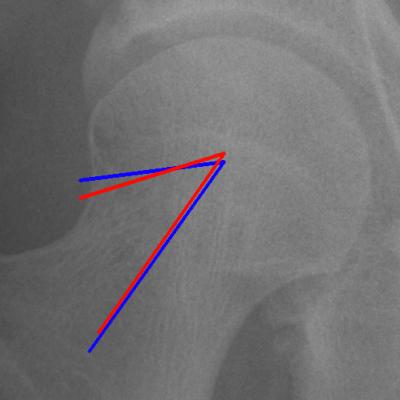
Adult hip x-ray — hip/joint angles
|
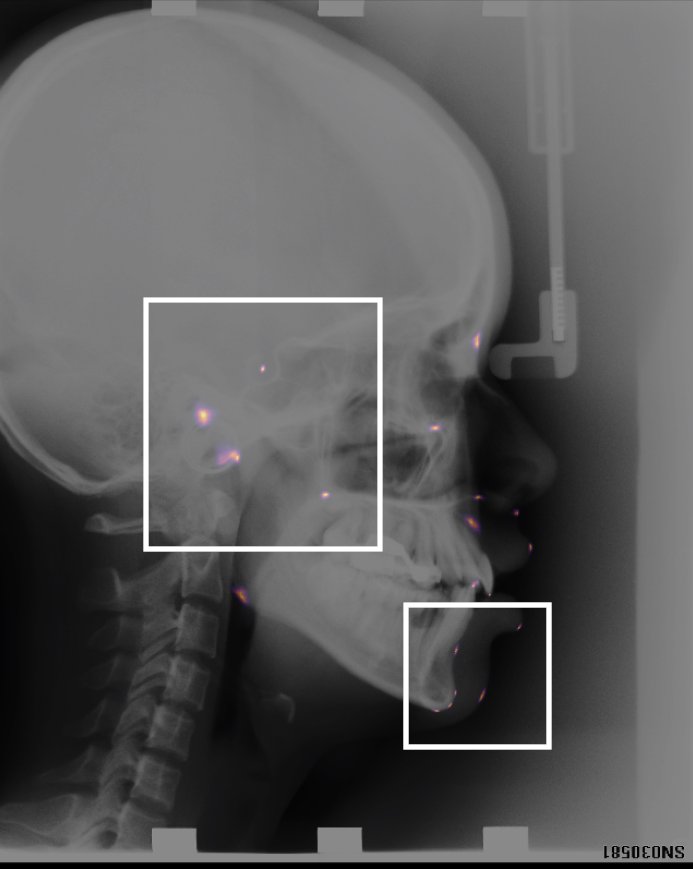
Landmark detection x-ray — cephalometric landmarks
|
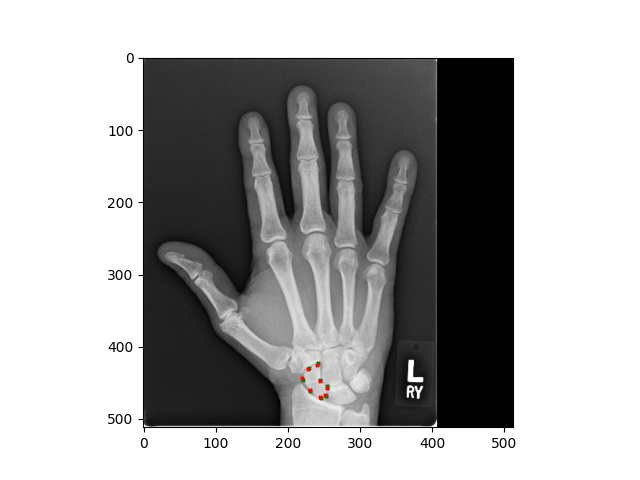
Adult hand x-ray — scaphoid bone
|
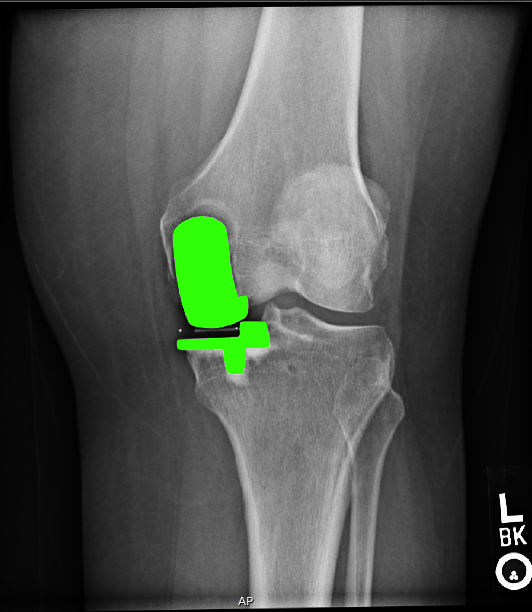
Unicompartmental knee x-ray
|
The key technology behind taking measurements relies on being able to identify anatomical landmarks. In our group we have designed bespoke landmark identification methods, as well as a novel way of displaying them. This work has been validated on a publicly available cephalometric dataset. We are also winners of the 2024 MICCAI landmark detection challenge.
We study hip x-rays to take angle measurements (such as the alpha angle) which are clinically relevant. Our results have been validated extensively on research datasets.
We detect features related to unicompartmental knee replacement and investigate clinical issues around fitting this kind of knee prosthesis.
Ultrasound
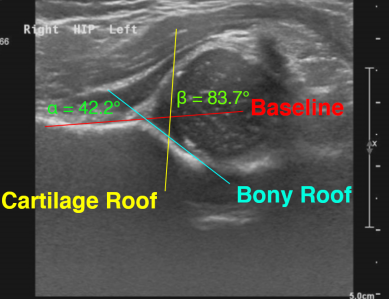
Infant DDH — developmental dysplasia angles
|
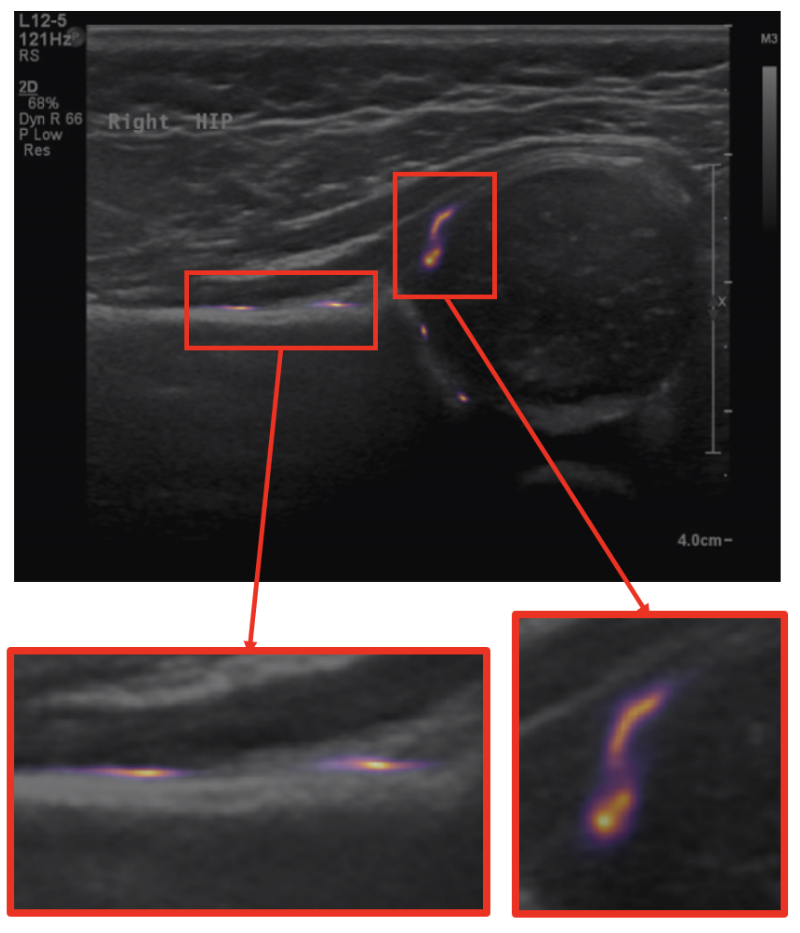
Measurements through landmark detection
|
Recent work has involved the study of ultrasound scans of infant hips, with a view to screening for developmental dysplasia of the hip (DDH). This work has scope for being translated into clinics.
MRI
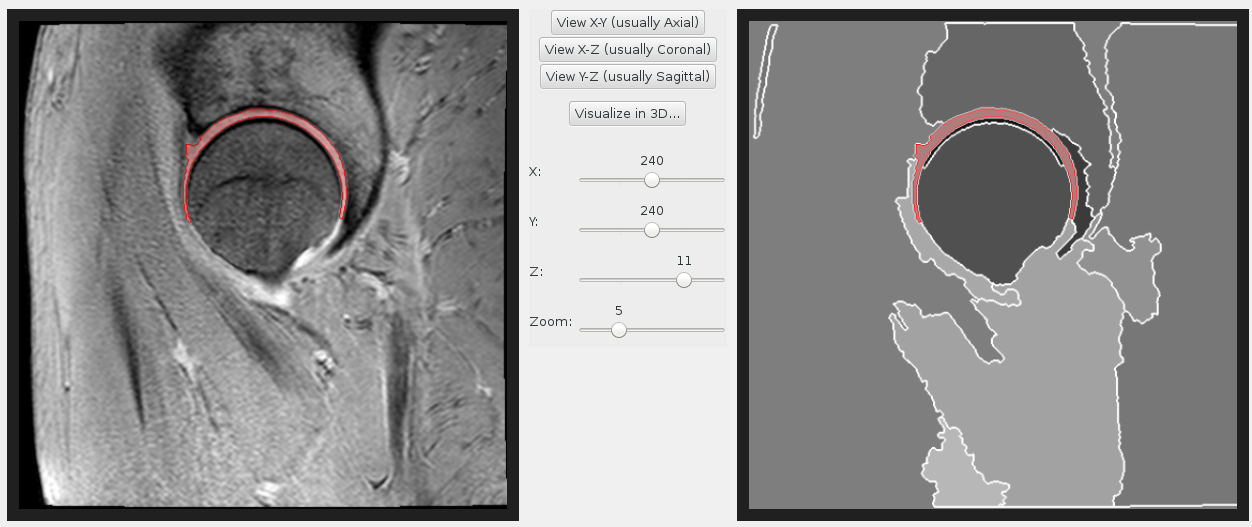
Hip cartilage MRI segmentation
|
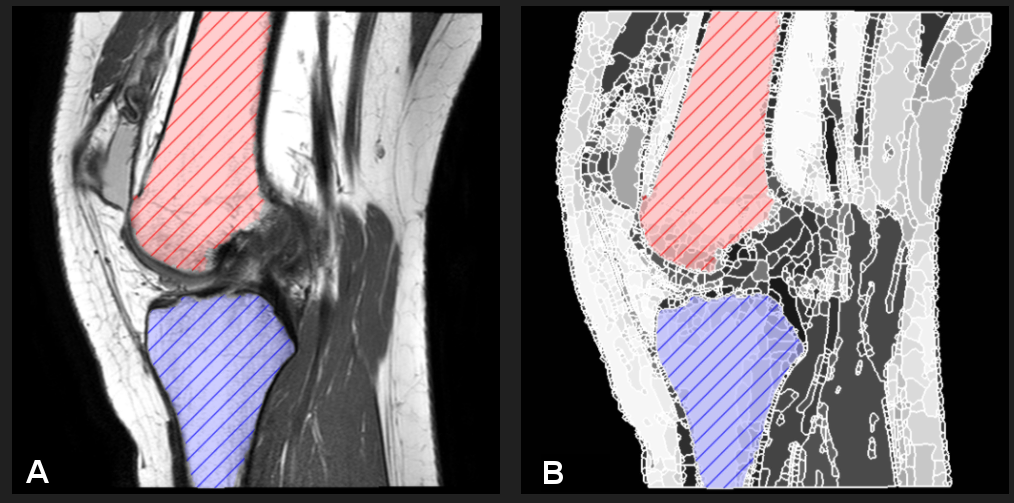
Knee MRI segmentation
|

BRATS brain MRI — tumour segmentation
|
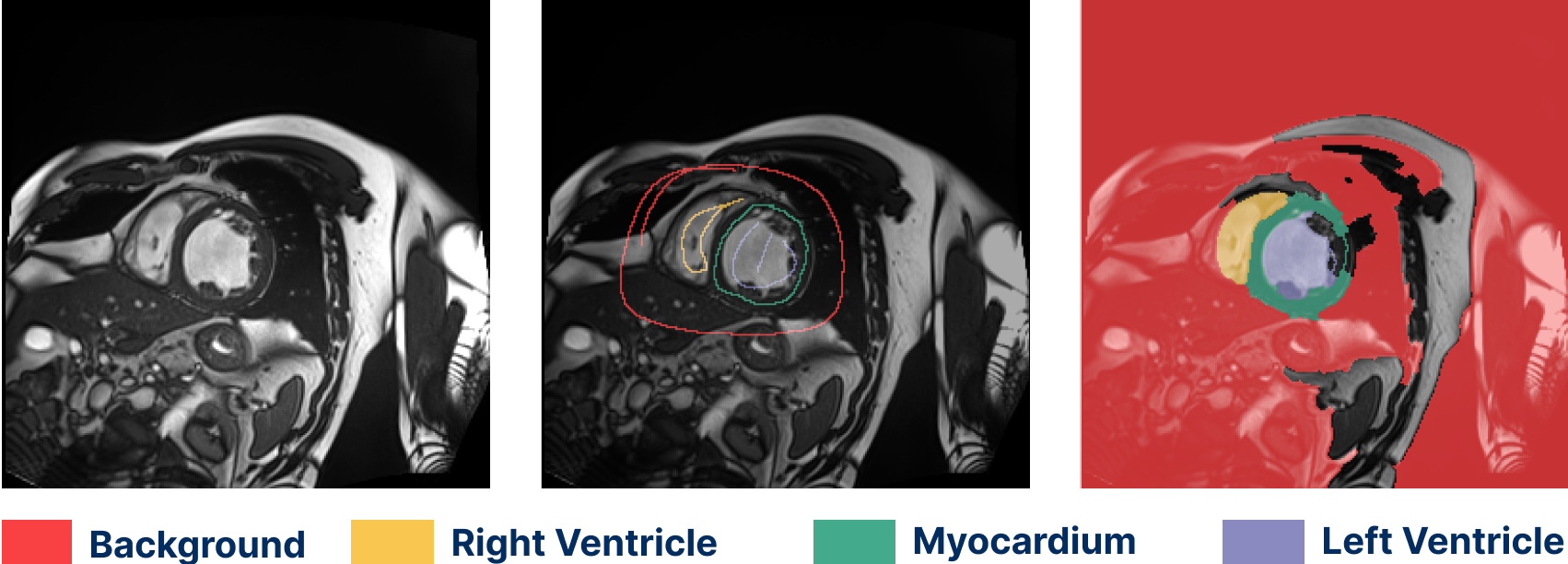
Cardiac MRI segmentation (ACDC dataset)
|
Semantic segmentation work has been both semi- and fully automated. Our methods have been used to label hip and knee joint bones, and also hip cartilage. We have extensive experience with a variety of MRI protocols. Segmentation can be fully supervised (from masks or contours) or weakly supervised (from scribbles).
CT
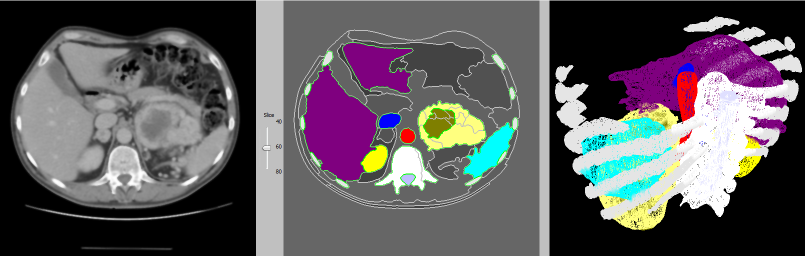
Abdominal CT — segmentation & 3D reconstruction
|
This 3D reconstruction of an abdominal CT scan features the rib cage, liver, kidneys, spleen, blood vessels and a renal tumour. Some of our techniques are capable of identifying kidneys automatically, as well as other organs in CT scans of the abdomen.
We detect features related to unicompartmental knee replacement and investigate clinical issues around fitting this kind of knee prosthesis.
Dermoscopy
We classify skin lesions into multiple, potentially overlapping classes. We calibrate our model to ensure that, when it is confident in its prediction, the prediction is also correct.
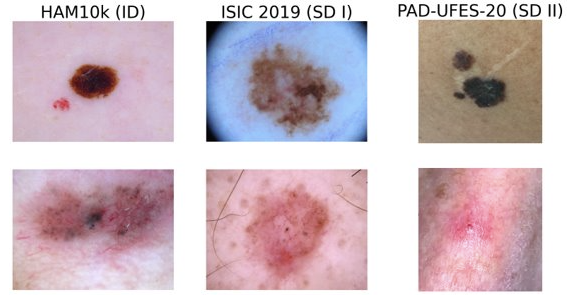
Dermatoscopic images — multi-class classification
|
Other interests
Over the years, my other interests have also included
- 3D modelling, 3D printing
- Multi-dimensional modelling
- Constructive Solid Geometry (CSG)
- Path planning
- Mathematics of curves and surfaces
- Finding roots of polynomials (for 3D modelling)
- Molecular modelling
Teaching
In the Geometric Modelling course we build interesting 3D models and design spline curves
Biography
I have been at the Department of Computer Science at Oxford since 1999. I obtained my PhD at the University of Bath, for research in Constructive Solid Geometry (CSG). I contributed to the development of the geometric modelling software svLis through the application of polynomial root finding methods.
As part of the Spatial Reasoning research group at Oxford I have been conducting research in the areas of molecular modelling (protein docking), medical imaging (CT and MRI scan analysis) and polynomial root finding (interval arithmetic). I am interested in other aspects of geometric modelling such as as applications of multivariate polynomial forms in geometry, Bernstein-form polynomials and generalized Newton-like methods.
I am actively involved in public engagement activities, such as the Oxford RobotGames, a programme where children of high school age design and build their own robot in half a day. Another on-going project of the Spatial Reasoning group has been to build a robot sheepdog which automatically herds either live or robotic animals. The Oxford robotic flock was shown in 2001 at the Royal Society Summer Science Exhibition.
I am Fellow of the UK Geometric Modelling Society and Associate Editor of the SPIE Journal of Medical Imaging.
Selected Publications
-
Automated landmark detection for assessing hip conditions: a cross−modality validation of MRI versus x−ray
Roberto Di Via‚ Vito Paolo Pastore‚ Francesca Odone‚ Sion Glyn−Jones and Irina Voiculescu
In International Symposium on Biomedical Imaging (ISBI). April, 2026.
Details about Automated landmark detection for assessing hip conditions: a cross−modality validation of MRI versus x−ray | BibTeX data for Automated landmark detection for assessing hip conditions: a cross−modality validation of MRI versus x−ray | Link to Automated landmark detection for assessing hip conditions: a cross−modality validation of MRI versus x−ray
-
Parallel Watershed Partitioning: GPU−Based Hierarchical Image Segmentation
Varduhi Yeghiazaryan‚ Yeva Gabrielyan and Irina Voiculescu
In Parallel and Distributed Computing (JPDC). November, 2025.
Details about Parallel Watershed Partitioning: GPU−Based Hierarchical Image Segmentation | BibTeX data for Parallel Watershed Partitioning: GPU−Based Hierarchical Image Segmentation | Link to Parallel Watershed Partitioning: GPU−Based Hierarchical Image Segmentation
-
Are X−ray landmark detection models fair? A preliminary assessment and mitigation strategy
Roberto Di Via‚ Massimiliano Ciranni‚ Davide Marinelli‚ Allison Clement‚ Nikil Patel‚ Julian Wyatt‚ Francesca Odone‚ Matteo Santacesaria‚ Irina Voiculescu and Vito Pastore
In International Conference on Computer Vision (ICCV). Systematic Trust in AI Models (STREAM) Workshop. October, 2025.
Details about Are X−ray landmark detection models fair? A preliminary assessment and mitigation strategy | BibTeX data for Are X−ray landmark detection models fair? A preliminary assessment and mitigation strategy | Link to Are X−ray landmark detection models fair? A preliminary assessment and mitigation strategy
