Oxford Medical Image Segmentation
The Oxford Medical Image Segmentation (OxMedIS) research group develops machine learning methods for medical image analysis, including segmentation, landmarks, classification and uncertainty.
Our group works across X-ray, MRI, CT, ultrasound, and other imaging data. We design algorithms that detect and classify clinically relevant features, reconstruct anatomical structures in 3D, and provide tools that balance automation with clinician input. Beyond image analysis, we address failure detection and model calibration to ensure reliability, with the ultimate goal of translating these methods into clinical practice.
Clinical Applications
Modalities
X-ray
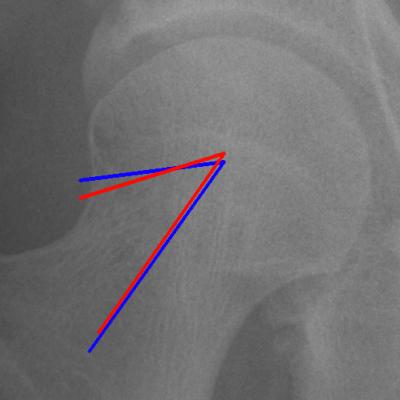 Adult hip x-ray — hip/joint angles
|
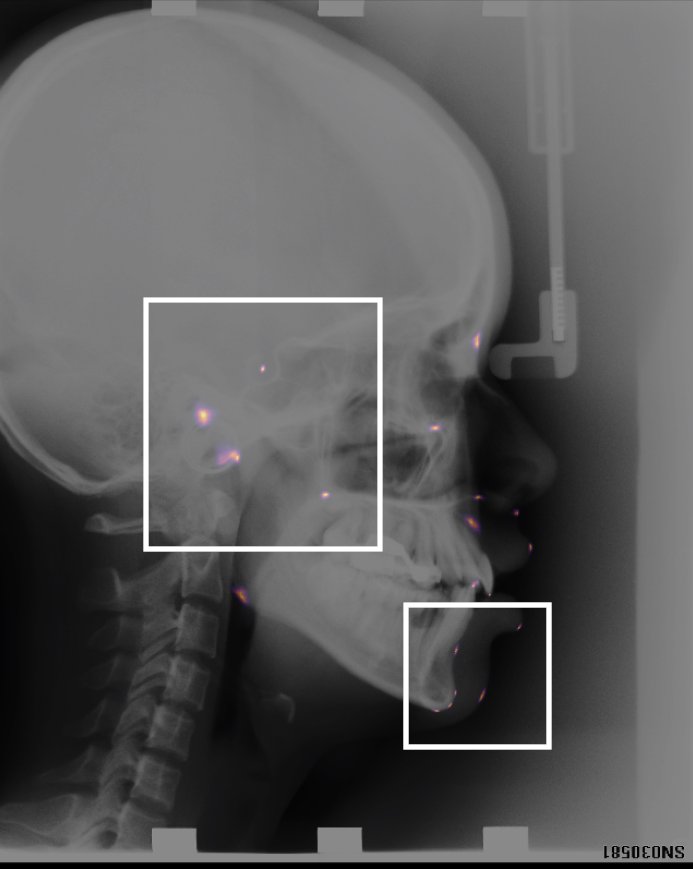 Landmark detection x-ray — cephalometric
|
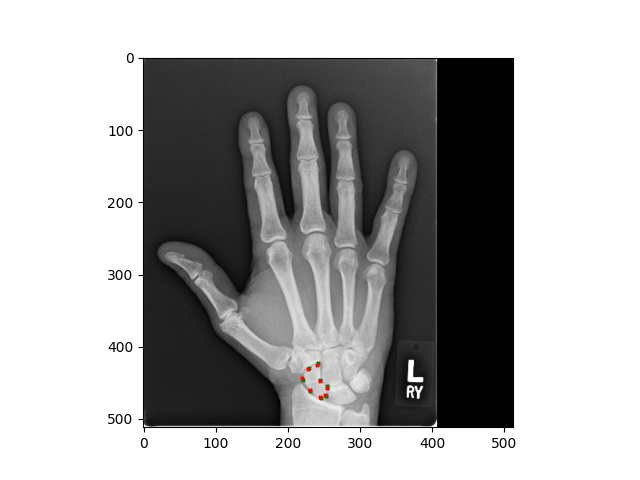 Adult hand x-ray — scaphoid bone
|
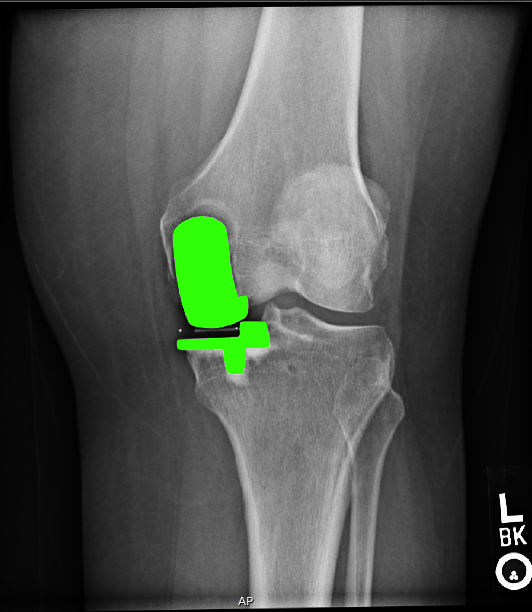 Unicompartmental knee x-ray
|
The key technology behind taking measurements relies on being able to identify anatomical landmarks. In our group we have designed bespoke landmark identification methods, as well as a novel way of displaying them. This work has been validated on a publicly available cephalometric dataset. We are also winners of the 2024 MICCAI landmark detection challenge.
We study hip x-rays to take angle measurements (such as the alpha angle) which are clinically relevant. Our results have been validated extensively on research datasets.
We detect features related to unicompartmental knee replacement and investigate clinical issues around fitting this kind of knee prosthesis.
Ultrasound
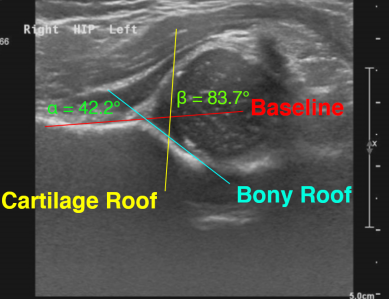 Infant DDH — developmental dysplasia angles
|
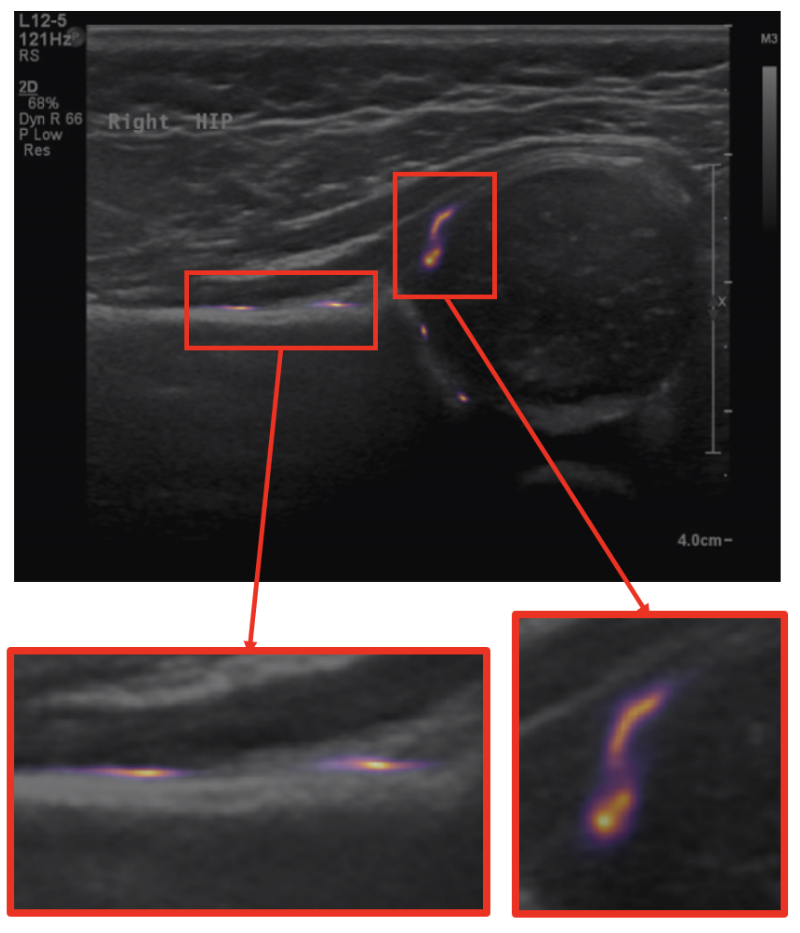 Measurements through landmark detection
|
Recent work has involved the study of ultrasound scans of infant hips, with a view to screening for developmental dysplasia of the hip (DDH). This work has scope for being translated into clinics.
MRI
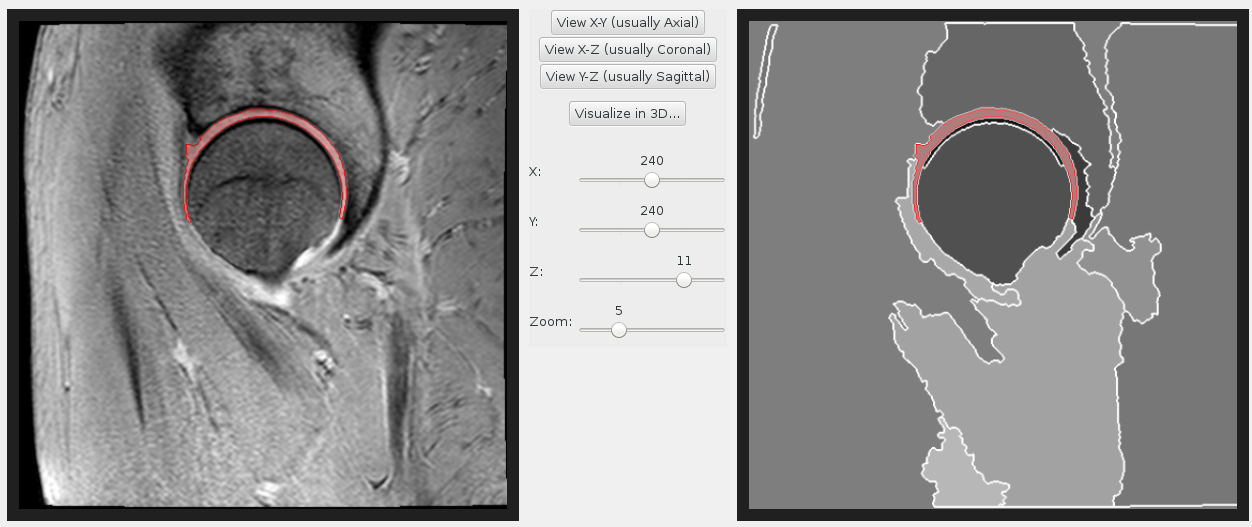 Hip cartilage MRI segmentation
|
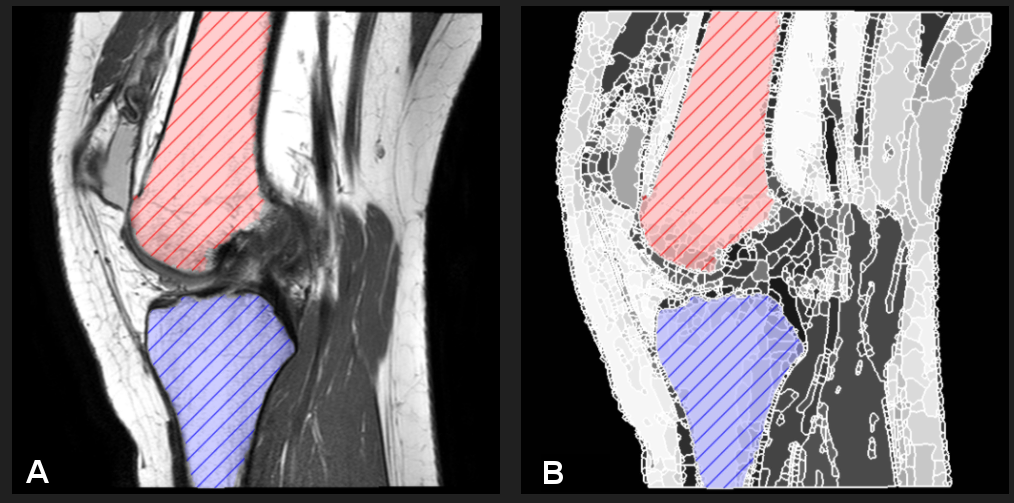 Knee MRI segmentation
|
 BRATS brain MRI — tumour segmentation
|
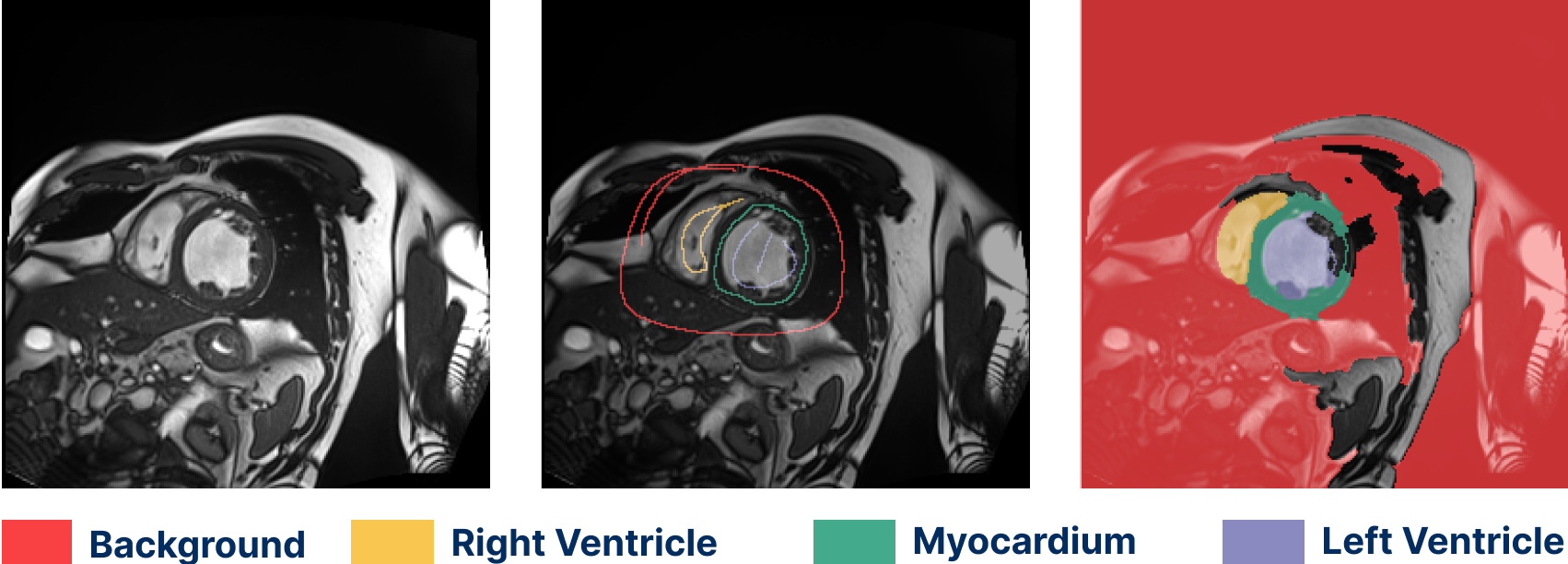 Cardiac MRI segmentation (ACDC dataset)
|
Semantic segmentation work has been both semi- and fully automated. Our methods have been used to label hip and knee joint bones, and also hip cartilage. We have extensive experience with a variety of MRI protocols. Segmentation can be fully supervised (from masks or contours) or weakly supervised (from scribbles).
CT
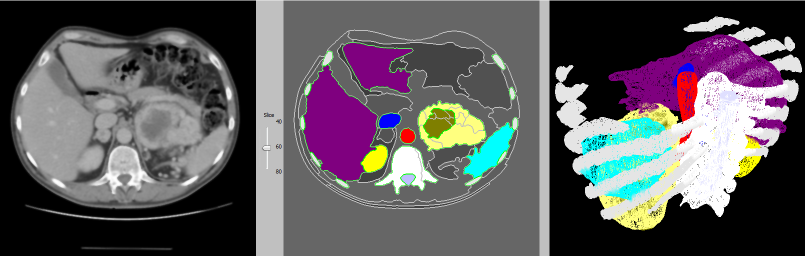 Abdominal CT — segmentation & 3D reconstruction
|
This 3D reconstruction of an abdominal CT scan features the rib cage, liver, kidneys, spleen, blood vessels and a renal tumour. Some of our techniques are capable of identifying kidneys automatically, as well as other organs in CT scans of the abdomen.
We detect features related to unicompartmental knee replacement and investigate clinical issues around fitting this kind
of knee prosthesis.
Dermoscopy
We classify skin lesions into multiple, potentially overlapping classes. We calibrate our model to ensure that, when it is confident in its prediction, the prediction is also correct.
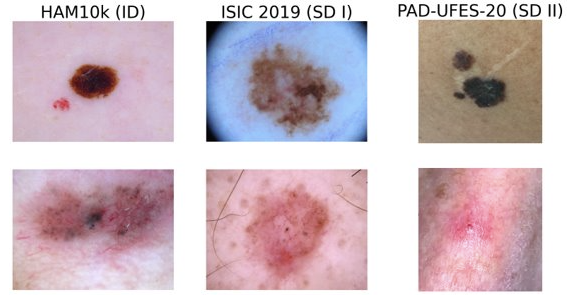 Dermatoscopic images — multi-class classification
|
Anatomy
We cover a range of different anatomical Specialities, such as:
- Musculoskeletal (hands, knees, hips, skull)
- Organ Systems (abdominal, cardiac, brain)
- Dermatology
- Histopathology
Please contact us for more information!
Technical Projects
Segmentation
We study the evaluation and development of automated segmentation methods.
Our recent work has focused on:
- Weakly supervised methods, such as those from scribbles or points.
- Improving network architecture for better performance (for example, loss functions).
- Evaluation and development of metrics.
Landmark Detection
We focus on enhancing landmark detection methods by:
- Improving multi-domain alignment (winner of the landmark detection challenge at MICCAI 2024!).
- Evaluating fairness.
- Developing few-shot detection.
Classification
We improve classification methods through:
- Network improvements, such as adding a clinical decision to the loss function.
- Developing techniques for reliable failure detection (Super-TrustScore)
Uncertainty and Confidence
Our most recent work develops novel methods for computing:
- Landmark uncertainty.
- Measurement confidence.
- Segmentation uncertainty.
